Big Picture
Because neural network comes with lots of mathematical notations (e.g. linear algebra, partial derivative sign
1. Initialize parameters (weights and biases) 2. Repeat the following until the loss becomes small i. With current parameters, compute predicted Y ii. Compute cost between actual Y and predicted Y iii. Compute the derivative of the cost with respect to parameters iv. Update current parameters by gradient descent 3. Predict by forward propagation with the updated parameters
People say the above i. step the forward propagation, and the iii. step the backward propagation. So I think we can simplify the above as followed.
Repeat this 1. Forward propagation 2. Compute cost 3. Backward propagation 4. Gradient descent
I oversimplied it because I wanted to emphasize that's the essence of the neural network, before we go to the details.
Forward Propagation
People use two type of parameters,
import math def sigmoid(x): return 1 / (1 + math.exp(-x)) def tanh(x): return (math.exp(x) - math.exp(-x)) / (math.exp(x) + math.exp(-x)) def relu(x): return max(0, x)
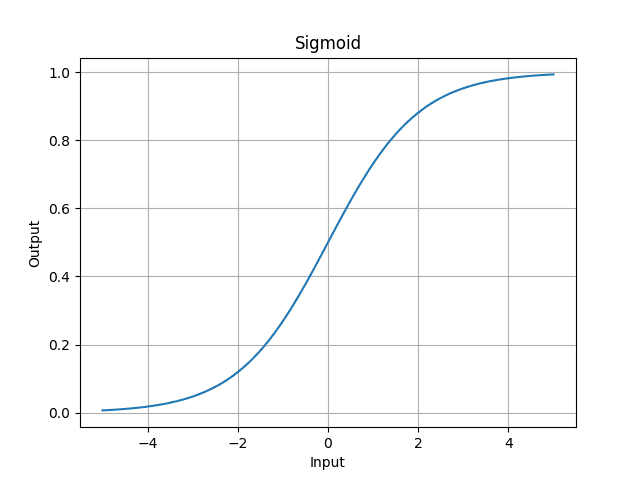
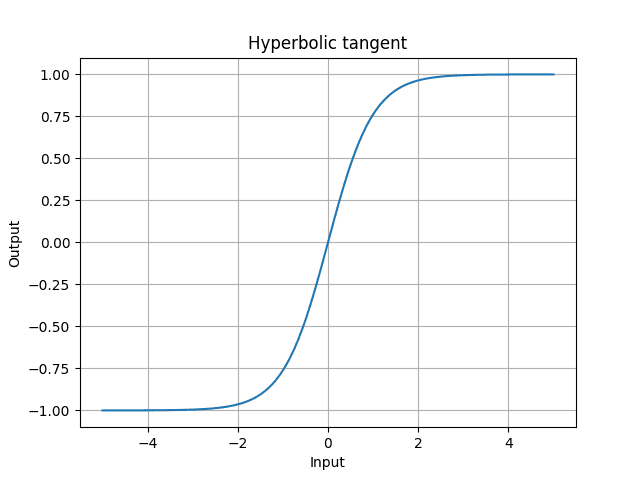
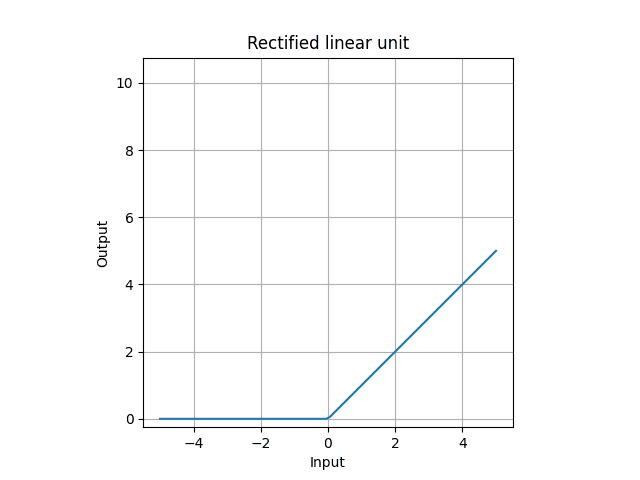
- sigmoid is something we want the output to be between 0 and 1.
- tanh is between -1 and 1
- relu is if input is positive, do nothing, but if negative, all 0.
In math, sigmoid is,
Tanh is,
ReLU is,
why we need to use non-linear activation. We see why by stacking 2 linear functions. Because we don't use non-linear activation and in practice it means don't use activation function at all to have linear activation,
By substituting the first equation to the second,
Let
This is a linear equation. So composition of linear functions is a linear function. If the data has a non-linearity, the neural network which only uses linear activations cannot express the relationship.
We go back to the previous linear equation and non-linear equation. In neural network, we are allowed to repeat it as many as we want. We can stack them many times. We can have as many layers as we want. Use
For example, the input layer is,
The output layer is,
Notice that when we have
- If we need to make binary classification,
needs to be a sigmoid function. - If we are asked to make regression,
is not a non-linear function. Just use the itself, because it will be a real number output computed by linear combination.
When a neural network has no hidden layers and one unit in output layer, meaning we are not repeating or stacking layers in neural network,
- If output layer is linear activation, it's linear regression because
, same as - If output layer is sigmoid activation, it's logistic regression because
where , same as
We check the dimension of weights
Bias is always one column, so to make the following calculation possible, broadcasting happens to bias in programming computation. For example,
In linear algebra, you cannot do element-wise addtion between
Notice that
Compute Cost
We define cost
We have
We will come back to the
Our goal is to minimize this cost function. In math,
It means that we want to minimize the cost function
Backward Propagation
In forward propagation, we sequentially computed from input layer with the given data
The reason why we have a sequence of derivative fractions is becuase
Backward propagation requires the derivative of the activation functions
Derivative of sigmoid is,
Derivative of hyperbolic tangent (tanh) is,
Derivative of rectified linear unit (ReLU) is,
Gradient Descent
Math behind Neural Network Binary Classification
I think the backward propagation part doesn't look clear unless we have a concrete neural network architecture. Here, we use ReLU for all the activation functions in the hidden layers, and use sigmoid for the output layer.
So this neural network does a binary classification. So the neural network architecture is,
Linear (
We skip the forward propagation part because it's only about doing linear combination of
In backward propagation, we start from the gradient in the output layer.
In backward propagation, the first derivative we need to compute is,
To make clear what's happening, instead of the cost
Derivative of loss function with respect to
So we have,
So we found that we just element-wise apply
What we need to compute next in backward propagation is,
To make things clear, we go back again to a single example case. notice that
By using the previously found result,
So we found that
Finally we can go to
By going back to a single example again, we have
By previously found result in the chain, the derivative of the loss with respect to weight is,
The derivative of the loss with respect to bias
In the entire data,
We have
We don't forget the gradient descent to update
For the next previous layer in backward propagation, we need to find
So the derivative with respect to
The derivative of the loss with respect to
Element-wise apply the above to
To continue the other layers in backward propagation, computation for the derivative with respect to
import numpy as np def relu(Z): A = np.maximum(0, Z) cache = Z return A, cache def relu_backward(dA, cache): Z = cache dZ = np.array(dA, copy=True) dZ[Z <= 0] = 0 return dZ def sigmoid(Z): A = 1 / (1 + np.exp(-Z)) cache = Z return A, cache def sigmoid_backward(dA, cache): Z = cache s = 1 / (1 + np.exp(-Z)) dZ = dA * s * (1 - s) return dZ def initialize_parameters(layer_dims, seed): """ Argument: layer_dims: List of integers representing dimenstions of each layer Return: parameters: Dictionary containing weight matrices for W (n_l, n_{l - 1}) and bias columns (n_l, 1) """ np.random.seed(seed) parameters = {} L = len(layer_dims) # Start from 1 to avoid index out of bound for l in range(1, L): # Initialize weights with small numbers so multiply by 0.01 parameters[f'W{l}'] = 0.01 * np.random.randn(layer_dims[l], layer_dims[l - 1]) # Initialize bias with zero parameters[f'b{l}'] = np.zeros((layer_dims[l], 1)) return parameters def linear_forward(A, W, b): Z = np.dot(W, A) + b cache = (A, W, b) return Z, cache def linear_activation_forward(A_prev, W, b, activation): Z, linear_cache = linear_forward(A_prev, W, b) if activation == 'relu': A, activation_cache = relu(Z) elif activation == 'sigmoid': A, activation_cache = sigmoid(Z) cache = (linear_cache, activation_cache) return A, cache def forward_propagation(X, parameters): caches = [] # A^0 = X A = X # // 2 because parameters are doubled by w and b L = len(parameters) // 2 # Hidden layers for l in range(1, L): A, cache = linear_activation_forward( A, parameters[f'W{l}'], parameters[f'b{l}'], # All the hidden layers use ReLU in this example activation='relu' ) caches.append(cache) # Output layer # AL = sigmoid(W^L A^{L - 1} + b^L) = \hat{Y} AL, cache = linear_activation_forward( A, parameters[f'W{L}'], parameters[f'b{L}'], # Use sigmoid activation because this example is binary classification activation='sigmoid' ) caches.append(cache) return AL, caches def compute_cost(AL, Y): """ Argument: AL: Output of sigmoid activation, so equal to \hat{y} Y: (1, number of examples) """ m = Y.shape[1] # Compute cross-entropy cost # np.multiply is element-wise multiplication cost = (-1 / m) * np.sum(np.multiply(Y, np.log(AL)) + np.multiply(1 - Y, np.log(1 - AL))) # np.squeeze removes all the length 1 dimensions, so [[1]] goes to 1, shape is () cost = np.squeeze(cost) return cost def linear_backward(dZ, cache): A_prev, W, b = cache m = A_prev.shape[1] dW = (1 / m) * np.dot(dZ, A_prev.T) db = (1 / m) * np.sum(dZ, axis=1, keepdims=True) dA_prev = np.dot(W.T, dZ) return dA_prev, dW, db def linear_activation_backward(dA, cache, activation): linear_cache, activation_cache = cache if activation == 'relu': dZ = relu_backward(dA, activation_cache) elif activation == 'sigmoid': dZ = sigmoid_backward(dA, activation_cache) dA_prev, dW, db = linear_backward(dZ, linear_cache) return dA_prev, dW, db def backward_propagation(AL, Y, caches): # Gradients grads = {} L = len(caches) m = AL.shape[1] # Make AL have the same shape as AL (Y hat) Y = Y.reshape(AL.shape) # Derivative of cost function with respect to A (sigmoid activation at the output layer) dAL = - (np.divide(Y, AL) - np.divide(1 - Y, 1 - AL)) current_cache = caches[-1] grads[f'dA{L - 1}'], grads[f'dW{L}'], grads[f'db{L}'] = linear_activation_backward( dAL, current_cache, 'sigmoid' ) # e.g. for i in reversed(range(3)): (2, 1, 0), # So it's (0, 1, 2) and then reverse it for l in reversed(range(L - 1)): current_cache = caches[l] grads[f'dA{l}'], grads[f'dW{l + 1}'], grads[f'db{l + 1}'] = linear_activation_backward( grads[f'dA{l + 1}'], current_cache, 'relu' ) return grads def update_parameters(parameters, grads, learning_rate): # // 2 because parameters are doubled by w and b L = len(parameters) // 2 for l in range(L): # Gradient descent parameters[f'W{l + 1}'] = parameters[f'W{l + 1}'] - learning_rate * grads[f'dW{l + 1}'] parameters[f'b{l + 1}'] = parameters[f'b{l + 1}'] - learning_rate * grads[f'db{l + 1}'] return parameters # For training # When input has 100 features, and to make 4 layers neural network # Last element needs to be 1 because of binary classification layer_dims = [100, 20, 7, 5, 1] learning_rate = 0.001 num_iterations = 1000 np.random.seed(0) # X: (p by n) X = np.array([]) # Y: (1 by n) Y = np.array([]) # Initialize parameters parameters = initialize_parameters(layer_dims) # Store cost computed from each iteration costs = [] # Keep training until num_iterations times for _ in range(num_iterations): # Forward propagation to compute Y hat AL, caches = forward_propagation(X, parameters) # Compute cost cost = compute_cost(AL, Y) # Backward propagation to compute gradients grads = backward_propagation(AL, Y, caches) # Update parameters by gradient descent parameters = update_parameters(parameters, grads, learning_rate) # Save cost costs.append(cost) # Visualize cost reduced over training iterations plt.plot(np.squeeze(costs)) plt.ylabel('cost') plt.xlabel('iterations') plt.title("Learning rate =" + str(learning_rate)) plt.show() # For predicting def predict(X, y, parameters): m = X.shape[1] n = len(parameters) // 2 p = np.zeros((1, m)) # Forward propagation to compute Y hat # In prediction, we don't need caches probas, _ = forward_propagation(X, parameters) # Convert the predicted probability to binary prediction for i in range(probas.shape[1]): if probas[0,i] > 0.5: p[0,i] = 1 else: p[0,i] = 0 print(f'Accuracy: {np.sum((p == y) / m)}') return p